VAE
Tags
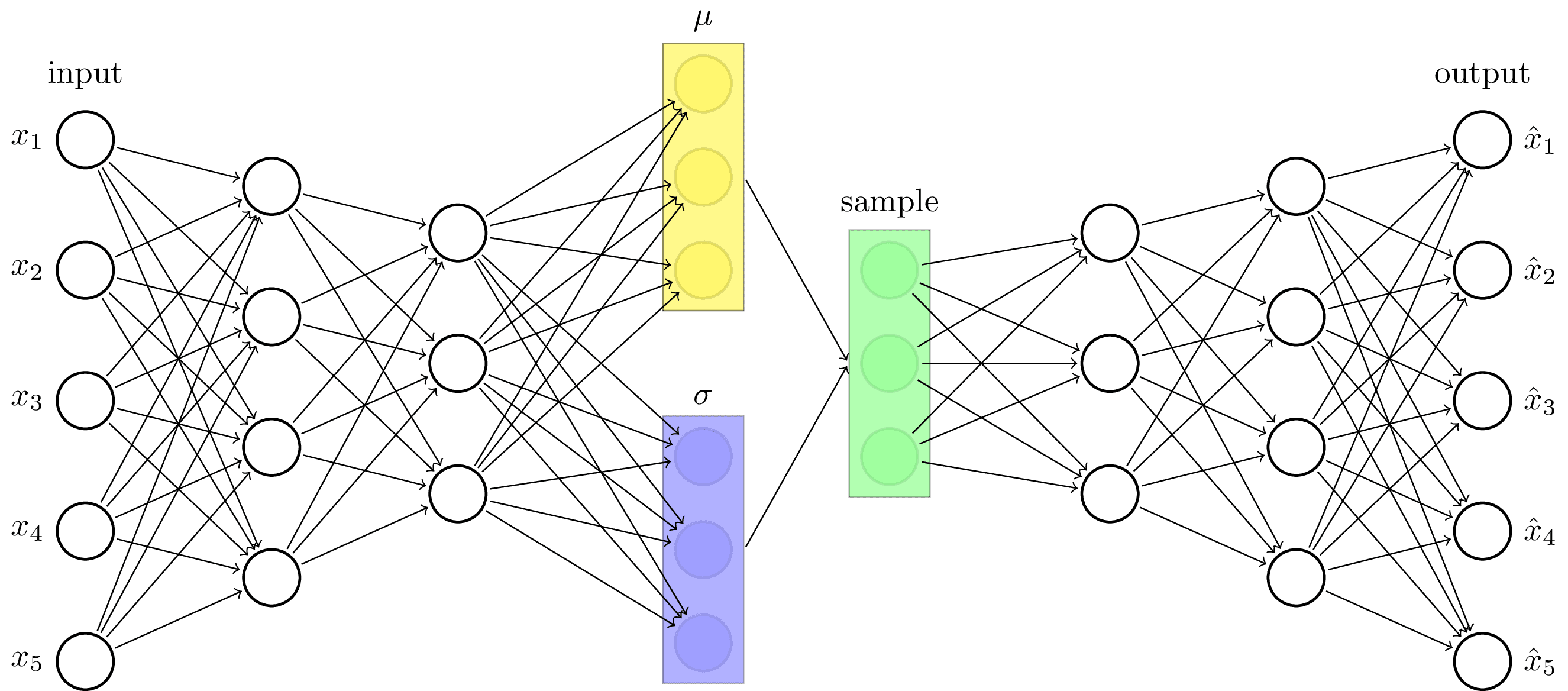
Variational autoencoder architecture. The earliest type of generative machine learning model. Inspired by https://towardsdatascience.com/intuitively-understanding-variational-autoencoders-1bfe67eb5daf.
Edit
Download
Code
vae.tex (78 lines)
\documentclass[tikz]{standalone}
\usepackage{xstring}
\usetikzlibrary{fit,positioning}
\newcommand\drawNodes[2]{
% #1 (str): namespace
% #2 (list[list[str]]): list of labels to print in the node of each neuron
\foreach \neurons [count=\lyrIdx] in #2 {
\StrCount{\neurons}{,}[\layerLen] % use xstring package to save each layer size into \layerLen macro
\foreach \n [count=\nIdx] in \neurons
\node[neuron] (#1-\lyrIdx-\nIdx) at (2*\lyrIdx, \layerLen/2-1.4*\nIdx) {\n};
}
}
\newcommand\denselyConnectNodes[2]{
% #1 (str): namespace
% #2 (list[int]): number of nodes in each layer
\foreach \n [count=\lyrIdx, remember=\lyrIdx as \previdx, remember=\n as \prevn] in #2 {
\foreach \y in {1,...,\n} {
\ifnum \lyrIdx > 1
\foreach \x in {1,...,\prevn}
\draw[->] (#1-\previdx-\x) -- (#1-\lyrIdx-\y);
\fi
}
}
}
\begin{document}
\begin{tikzpicture}[
shorten >=1pt, shorten <=1pt,
neuron/.style={circle, draw, minimum size=4ex, thick},
legend/.style={font=\large\bfseries},
]
% encoder
\drawNodes{encoder}{{{,,,,}, {,,,}, {,,}}}
\denselyConnectNodes{encoder}{{5, 4, 3}}
% decoder
\begin{scope}[xshift=11cm]
\drawNodes{decoder}{{{,,}, {,,,}, {,,,,}}}
\denselyConnectNodes{decoder}{{3, 4, 5}}
\end{scope}
% mu, sigma, sample nodes
\foreach \idx in {1,...,3} {
\coordinate[neuron, right=2 of encoder-3-2, yshift=\idx cm,, fill=yellow, fill opacity=0.2] (mu-\idx);
\coordinate[neuron, right=2 of encoder-3-2, yshift=-\idx cm, fill=blue, fill opacity=0.1] (sigma-\idx);
\coordinate[neuron, right=4 of encoder-3-2, yshift=\idx cm-2cm, fill=green, fill opacity=0.1] (sample-\idx);
}
% mu, sigma, sample boxes
\node [label=$\mu$, fit=(mu-1) (mu-3), draw, fill=yellow, opacity=0.45] (mu) {};
\node [label=$\sigma$, fit=(sigma-1) (sigma-3), draw, fill=blue, opacity=0.3] (sigma) {};
\node [label=sample, fit=(sample-1) (sample-3), draw, fill=green, opacity=0.3] (sample) {};
% mu, sigma, sample connections
\draw[->] (mu.east) edge (sample.west) (sigma.east) -- (sample.west);
\foreach \a in {1,2,3}
\foreach \b in {1,2,3} {
\draw[->] (encoder-3-\a) -- (mu-\b);
\draw[->] (encoder-3-\a) -- (sigma-\b);
\draw[->] (sample-\a) -- (decoder-1-\b);
}
% input + output labels
\foreach \idx in {1,...,5} {
\node[left=0 of encoder-1-\idx] {$x_\idx$};
\node[right=0 of decoder-3-\idx] {$\hat x_\idx$};
}
\node[above=0.1 of encoder-1-1] {input};
\node[above=0.1 of decoder-3-1] {output};
\end{tikzpicture}
\end{document}